Acute hepatitis B: national enhanced surveillance report January to December 2022
Updated 24 January 2024
Background
The quarterly reporting of enhanced molecular surveillance of acute hepatitis B is based on clinical reports of acute cases to UK Health Security Agency (UKHSA) being entered on HPZone and corresponding samples being submitted to the UKHSA Blood Borne Virus Unit (BBVU) in the Virus Reference Department (VRD) at Colindale.
In 2016, VRD re-introduced anti hepatitis B core avidity testing alongside genotyping of samples from patients diagnosed with acute hepatitis B, a service that is offered free of charge. Hospital microbiology and virology departments are requested to send samples to Colindale for confirmation, avidity testing and genotyping as part of the national enhanced surveillance of acute hepatitis B (see Acute hepatitis B: guide to national enhanced surveillance).
Following the reporting of clusters of acute hepatitis B in 2016, an HPZone Context ‘Acute hepatitis B’ was added for monitoring of acute cases.
Methods
Records of an ‘Acute Hepatitis B’ reported between January and December 2022 were collated from HPZone, and through samples submitted to the UKHSA BBVU at Colindale.
Acute hepatitis B cases are recorded on HPZone in 2 different ways:
- HPZone Context ‘Acute Hepatitis B’ data includes personally identifiable information, which therefore allows for the rapid identification of cases and the requesting of samples directly from laboratories for avidity and molecular characterisation at VRD Colindale
- HPZone Dashboard data contains no personally identifiable information but can be linked to Context data using the HPZone unique identifier
The ‘Acute Hepatitis B’ Context data was matched to laboratory testing data from VRD using a combination of Surname, First name, Date of birth, Sex, and NHS number.
Results
Between January and December 2022, 213 cases of acute hepatitis B were reported onto HPZone Dashboard across England (confirmed, probable and possible).
Overall, the cases entered on HPZone Dashboard have been declining since 2012: from 485 in that year to 201 in 2021. Monthly cases since 2010 in England are shown in Figure 1. In 2015 there was a slight increase in cases likely caused by the outbreak of acute hepatitis B in men who have sex with men (MSM) who identify as heterosexual (1).
Figure 1. Cumulative cases of acute hepatitis B in England entered on HPZone Dashboard: 2010 to December 2022 (note 1)

Note 1: 2022 data is provisional
Figure 2. January to December 2022 cases entered onto HPZone Dashboard
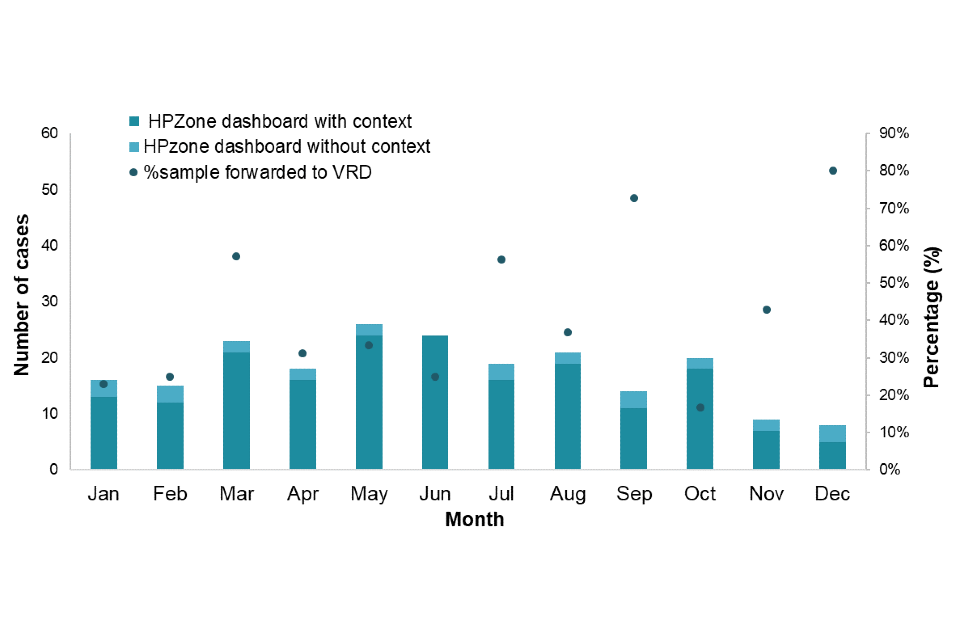
Note 2: The dots (right axis) show the proportion of HPZone Context cases that had a sample forwarded to VRD
Figure 2 shows the number of cases reported with personal identifiable information through HPZone Context by month. The additional information allows for a letter to be sent to request residual samples directly from laboratories for avidity and molecular characterisation. The dots represent the proportion of samples received from laboratories. Figure 3 shows this distribution by region.
Figure 3. January to December 2022 cases entered onto HPZone Context and/or entered onto HPZone Dashboard by UKHSA regions
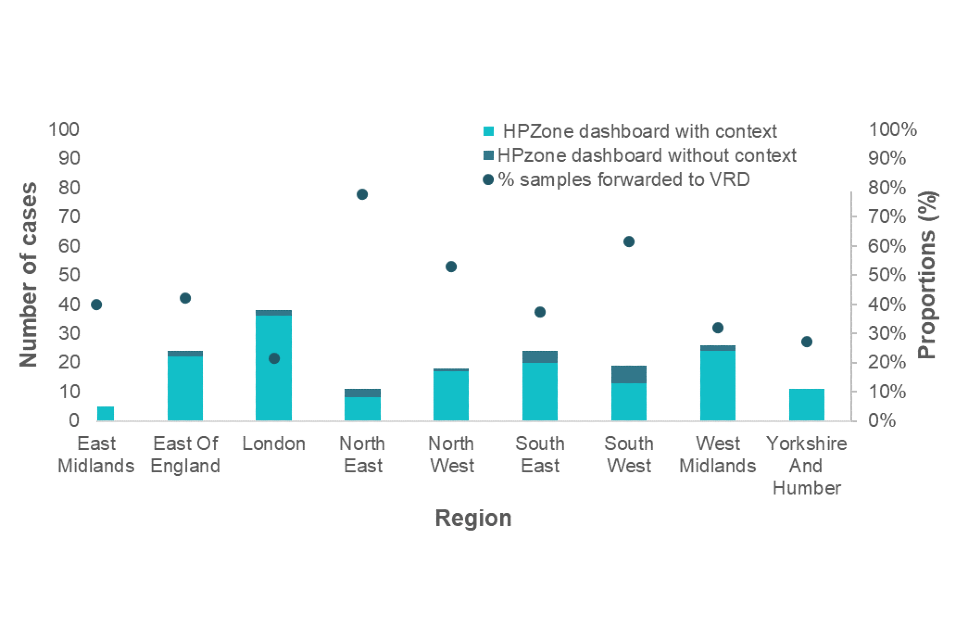
Note 3: The dots (right axis) show the proportion of HPZone Context cases that had a sample forwarded to VRD
For cases reported between January and December 2022, in both the HPZone Context dataset and the HPZone Dashboard dataset, age and sex were well reported (99%). Where sex was known , males accounted for 60% of cases (116 out of 193), and females 40% (77 out of 193). The median age of those with acute hepatitis B virus (HBV) was 46 years (interquartile range (IQR): 31 to 57): 49 (IQR: 33 to 59) for males and 40 (IQR: 29 to 55) for females.
The age distribution by sex is presented in Table 1. Overall, the highest proportion of cases was seen in the 45-to-54-years age group, followed by the 25-to-34-years age group. The highest proportion in males was seen in the 45-to-54-years age group, while in females it was in the 35-to-44-years age group.
Fifty-one percent (35 out of 69) of those with acute HBV were resident in the 2 most deprived quintiles. Where ethnicity was available, 55% (60 cases) of those with acute HBV (33 cases) were of white British ethnic origin, followed by those of Asian or Asian British ethnic origin (18%; 11 cases).
Table 1. Number and proportion of acute HBV cases from HPZone Dashboard by sex and age group during January to December 2022
| Age group | Female | Male | Unknown | Total |
|---|---|---|---|---|
| Under 15 | 0 (-) | 2 (2%) | 1 (50%) | 3 (2%) |
| 15 to 24 | 10 (13%) | 6 (5%) | 1 (50%) | 17 (9%) |
| 25 to 34 | 17 (22%) | 24 (21%) | 0 (-) | 41 (21%) |
| 35 to 44 | 19 (25%) | 13 (11%) | 0 (-) | 32 (16%) |
| 45 to 54 | 11 (14%) | 31 (27%) | 0 (-) | 42 (22%) |
| 55 to 64 | 12 (16%) | 19 (16%) | 0 (-) | 31 (16%) |
| 65 and over | 8 (10%) | 21 (18%) | 0 (-) | 29 (15%) |
| Total | 77 | 116 | 2 | 195 |
Avidity testing and molecular characterisation investigations were undertaken on samples linked to cases to confirm the acute hepatitis B diagnosis with additional genotyping and phylogenetic analysis to inform on the diversity of the circulating viruses.
Of the 98 samples submitted to VRD as part of the enhanced surveillance programme, 15 samples were confirmed to be from individuals with chronic hepatitis B and 34 were confirmed to be from individuals with acute hepatitis B infection. The avidity testing in 19 samples was classified as undetermined where it was not possible to confidently assign an HBV infection status. Thirty samples were not tested.
Not all cases with samples forwarded to VRD could be matched to cases in HPZone Context. This could either be due to a case not being entered on HPZone or it could be due to the case being entered in a previous quarter.
A total of 32 out of 34 confirmed acute cases could be genotyped during the January-to-December-2022 period. The distribution of genotypes is shown in Table 2. Consistent with trends seen in previous years, genotype A was the most commonly reported genotype, accounting for 38% of cases. Additional sub-genotype analysis of the A viruses indicated the majority to be A2 with only one case reported as A1. The distribution of genotypes seen in UKHSA regions is shown in Figure 4.
Table 2. Genotype distribution and proportions of acute hepatitis B cases tested at VRD in January to December 2022
| Acute genotype | Number of cases | Proportion of cases |
|---|---|---|
| A (note4) | 12 | 38% |
| B (note 4) | 2 | 6% |
| C (note 4) | 4 | 13% |
| D (note 4) | 9 | 28% |
| E | 5 | 16% |
| F | 0 | – |
| Total | 32 | – |
Note 4: Total excludes 2 DNA negative. Sub-genotype breakdowns were: A1=1 and A2=11; B2=1, B4=1; C1=2 and C2=2; D1=8 and D3=1.
Figure 4. Genotypes of acute samples sent to VRD by UKHSA region (note 5)
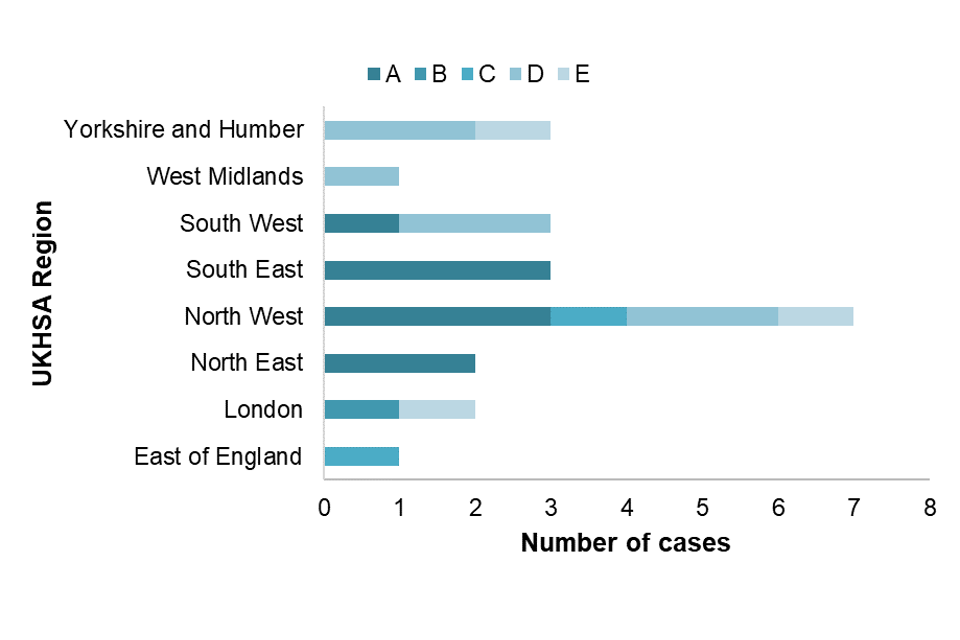
Note 5: Excludes 9 where region was unknown (genotypes A=3, C=2, E=2 and 2 cases where DNA was reported negative).
Discussion
Quarterly publication of enhanced molecular surveillance using matched HPZone and reference laboratory confirmatory and typing data with a regional breakdown allows real-time monitoring of acute hepatitis B transmission. The number of acute hepatitis B cases in January to December 2021 remained low and consistent with annual trends for the same timeframe. Molecular analysis provides insight into the current hepatitis B genotypes circulating in England, although interpretation is limited by the small proportion of samples submitted to VRD. The A2 ‘prisoner variant’ is one of the most common strains and is known to be well-established in the UK MSM population.
Other genotypes can indicate a geographical origin which can help provide an understanding of sources of infection and transmission routes. For example, genotype D is associated with South Asia. Timely assignment of cases to the HPZone Context and improved submission of samples for molecular characterisation will allow for more comprehensive monitoring of acute hepatitis B infection in England.
Reference
- Shankar AG, Mandal S, Ijaz S. An outbreak of hepatitis B in men who have sex with men but identify as heterosexual British Medical Journal Sexually Transmitted Infections 2016: volume 92, issue 3, page 227
