DNA relationship testing using autosomal short tandem repeats (accessible)
Updated 22 July 2024
FSR-G-228
Issue 1
1. Introduction
1.1 Background
1.1.1 The question of how individuals are related to each other arises in various contexts in criminal cases, as shown in the following examples.
a. It may be necessary to establish the identity of a body using reference DNA samples from blood relatives of the deceased.
b. Following a rape allegation that has led to the conception of a child, the question of its paternity will be material evidence in establishing that an act of sexual intercourse took place between the parties (this would not address the issue of consent).
c. If an alleged rape occurs between parties who are themselves biologically related, then the child will have been ‘incestuously’ conceived. In such instances, there may therefore be an important additional evidential element to establish that the biological parents are themselves related in some way (as well as being the biological parents of the child).
d. A case may be brought against a couple who are in a consensual sexual relationship but who are believed to be biologically related in such a way that makes that relationship a criminal offence (incest).
e. In human trafficking cases, individuals may assert that a child in their
f. custody is biologically theirs.
1.1.2 In whatever context it arises, it is important that forensic practitioners providing DNA relationship testing evidence to the criminal courts adhere to the same standards and principles as the practitioners presenting ‘traditional’ DNA evidence regarding identity.
1.1.3 However, it is important to recognise that using DNA to test biological relationships is a very different type of work to identity testing, and so it merits separate standards and guidance. This document is intended to fill that gap.
2. Other standards for DNA profile interpretation
2.1.1 National and international standards for testing and calibration in laboratories provide information on analytical methods used in forensic science and this document references those resources when applicable.
2.1.2 This document should be read alongside other documents published by the Forensic Science Regulator that impinge on these standards and guidance (see section 3).
2.1.3 Any guidelines produced in this document are, wherever possible, based on published work. These publications are referenced where relevant.
2.1.4 The content of this document has also been the subject of widespread consultation with practitioner representatives of all of the major providers of forensic science services in the UK and Ireland via the DNA Analysis Specialist Group.
3. Other applicable forensic science regulator codes and guidance
3.1.1 For England and Wales, as a minimum the requirements set out in the Forensic Science Regulator’s Codes of Practice and Conduct for Forensic Science Providers and Practitioners in the Criminal Justice System, FSR-C- 100, (the Codes) and FSR-C-108 DNA Analysis shall apply equally to relationship testing.
3.1.2 The relevant documents to this topic are:
c. FSR-G-202 The interpretation of DNA evidence (including low-template DNA);
f. FSR-G-222 DNA mixture interpretation;
g. FSR-G-223 Software validation for DNA mixture interpretation; and
h. FSR-P-302 DNA contamination detection: The management and use of staff elimination DNA databases.
4. Scope
4.1.1 The purpose of this document is to set standards and provide guidance for those forensic practitioners providing relationship testing services using autosomal DNA short tandem repeat (STR) profiling to the criminal justice system (CJS).
4.1.2 This document applies only to STR markers (although the ethos of the guidelines can be applied to other classes of autosomal markers such as In/Dels and single nucleotide polymorphisms (SNPs).
4.1.3 These standards and guidelines apply to courts operating at any level within the CJS of England and Wales.
4.2 Exclusions
4.2.1 Testing using uniparentally inherited markers located on the sex chromosomes and mitochondrial DNA is specifically excluded.
4.2.2 For the avoidance of doubt, the Forensic Science Regulator’s remit in requiring compliance to these standards and guidelines does not extend to:
a. Disaster victim identification (DVI) other than where there is a requirement to identify the remains of a suspected perpetrator;
b. Inquests held before HM Coroners of England and Wales in which evidence is adduced regarding the identity of a deceased person based on an analysis of the DNA of his/her putative relatives;
c. Clinical applications such as in utero paternity testing and other forms of genetic testing for medical or research purposes;
d. Family law matters such as child custody and maintenance arrangements;
e. Civil law matters such as inheritance disputes;
f. Immigration matters such as family repatriation; and
g. Privately commissioned voluntary tests (so-called ‘peace of mind’ tests).
4.2.3 For those areas of relationship testing where these standards and guidelines do not officially apply, nonetheless, the principles of good practice espoused herein may still be applied to those spheres of work.
5. Implementation
5.1.1 This appendix is available for incorporation into a forensic science provider’s (FSP’s) quality management system from the date of publication. The Regulator required that the Codes were included in a FSP’s schedule of accreditation from October 2017 and the requirements in this appendix are effective from 01 October 2021.
6. Modification
6.1.1 This is the first issue of this document.
6.1.2 The Regulator uses an identification system for all documents. In the normal sequence of documents this identifier is of the form ‘FSR-#-###’ where (a) (the first ‘#’) indicates a letter to describe the type of document and (b) ‘###’ indicates a numerical, or alphanumerical code to identify the document. For example, this document is FSR-G-228, and the ‘G’ indicates that it is a guidance document. Combined with the issue number this ensures that each document is uniquely identified.
6.1.3 If it is necessary to publish a modified version of a document (for example, a version in a different language), then the modified version will have an additional letter at the end of the unique identifier. The identifier thus becoming FSR-#-####.
6.1.4 In all cases the normal document bearing the identifier FSR-#-### is to be taken as the definitive version. In the event of any discrepancy between the normal version and a modified version then the text of the normal version shall prevail.
7. Terms and definitions
7.1.1 The word ‘shall’ has been used in this document where there is a corresponding requirement in ISO/IEC 17025:2017, the Forensic Science Regulator’s Codes or there is a corresponding legal requirement.
7.1.2 The word ‘should’ has been used to indicate generally accepted practice where the reason for not complying or any deviation shall be recorded.
7.1.3 The word ‘may’ or ‘might’ or ‘can’ or similar words indicates a voluntary component in the guidance.
7.1.4 The terms and definitions set out in the Forensic Science Regulator’s Codes, FSR-C-108 DNA Analysis, FSR-G-202 The Interpretation of DNA evidence (including low-template DNA), FSR -G-213 Allele frequency databases and reporting guidance for the DNA (short tandem repeat) profiling and FSR-G-222 DNA mixture interpretation apply to this document.
7.1.5 For the avoidance of doubt, the phrases ‘kinship testing’ and ‘pedigree testing’ are taken to be synonymous with ‘relationship testing’ and for consistency, this document will use the phrase ‘relationship testing’ as a generic term to cover all potential biological relationships between individuals.
7.1.6 In this document, if the term ‘paternity testing’ is used, it is restricted to its specific meaning (for example, investigating an alleged father/child relationship).
8. Quality control
8.1 Laboratory processing
8.1.1 The laboratory processing the samples and preparing the DNA profiles shall be accredited to ISO17025. The requirements set out in FSR-C-108 DNA Analysis shall be complied with.
8.2 Chain of custody
8.2.1 If it is anticipated that the evidence produced is to be used within the criminal justice system then the samples required for testing shall follow a chain of custody from sampling to presenting the evidence in court. This includes unique labelling of exhibits, appropriate documentation as well as tracking/recording of the movement of items.
8.2.2 For any samples taken outside the police chain of custody, the handling, transportation and receipt of such samples should nevertheless be documented.
8.3 Formalities for taking DNA samples for relationship testing
8.3.1 The identity of an individual voluntarily providing a DNA sample shall be established by the person requesting that sample:
a. Where relevant, a photograph of the person (or a copy of photographic identification they have supplied as proof of identity); or
b. A determination by an official authority on identity may be made to assist in proving identity should this later become an issue in the case.
8.3.2 Informed consent for the taking, use and retention of a voluntary sample (and the information derived from it) shall be obtained before the sample is taken. The volunteer, or an appropriate adult, shall specifically consent to the use of the sample for relationship testing purposes in order to proceed. This consent shall be documented accordingly. A sampling kit containing a central form[footnote 1] is specifically provided for this purpose.
8.3.3 If the person is deceased, then a DNA sample shall only be taken under the lawful authority of HM Coroners of England and Wales or pursuant to police powers of seizure.
8.3.4 A reference sample shall only be taken coercively from an arrested person using the powers granted to the police by virtue of the Police and Criminal Evidence Act 1984 (PACE). A sampling kit containing a central form[footnote 2] is specifically provided for this purpose.
8.3.5 Samples taken coercively pursuant to PACE shall only be used for only a limited set of purposes (which are set out in the Act) and which include the prosecution of crime and identification of the dead (i.e. for use in criminal and coronial cases). Such samples (which include the information derived from those samples) cannot therefore, by law, be used for a collateral purpose, such as a family or civil case, see Home Office Circular 1/2006.
8.3.6 Resampling and retesting shall therefore be required if civil or family matters follow on from a criminal case.
8.4 Guidance as to sample types suitable for relationship analysis
8.4.1 The type of sample available for relationship testing will depend on the circumstances of the individual case.
8.4.2 For living individuals, the usual sample type may be buccal (mouth) swabs. However, if the individual is a young child or an adult is undergoing medical treatment, then saliva, blood or freshly pulled hairs (with roots) might be an appropriate alternative. With neonates, blood from a heel stab (such as a Guthrie card) may be used, provided that the sample is traceable to the child via medical records.
8.4.3 For deceased individuals any available post-mortem sample can be utilised. If the cadaver is in an advanced state of decay, testing of skeletal structures such as bones and teeth may be required.
8.4.4 For a child in utero, following either a termination of a pregnancy or following a miscarriage, the police may use their powers of seizure to take possession of the products of conception, see Faculty of Forensic and Legal Medicine so that DNA samples from the foetus may be secured.
8.4.5 For missing individuals or to identify the deceased a ‘surrogate’ reference sample could be required. Such samples may be obtained from:
a. A traceable medical archive sample (such as wax embedded tissue, serum samples, cervical smears or other preparations on microscope slides); or
b. A personal effect attributable to the missing person or the deceased (preferably an item likely to have been used exclusively by that person and no one else) such as a toothbrush, dentures, a razor or a hairbrush.
8.4.6 When identifying a perpetrator’s remains in disaster victim identification (DVI) incidents, further guidance is provided in Interpol, Disaster Victim Identification (DVI) Protocols and Standards, Interpol, Best Practice Principles: Recommendations on the Use of DNA for the Identification of Missing Persons and Unidentified Human Remains, the College of Policing authorised professional practice, and the International Society for Forensic Genetics (ISFG) recommendations in Prinz et al., 2007.
8.5 Packaging
8.5.1 The packaging of collected biological material for forensic examination shall be fit for purpose. Specifically, the packaging shall:
a. Preserve the integrity of the material;
b. Minimise the risk of loss and/or degradation of the material;
c. Prevent adulteration or contamination of the material; and
d. Prevent the escape of any biohazardous material from the sample.
8.5.2 As a minimum this should include:
a. Separate packaging of items where the packaging of items together could compromise them;
b. Appropriate packaging for the size, condition and forensic analysis requirements of the material recovered;
c. Secure and tamper-evident seals; and
d. Where liquids are to be stored frozen, suitable containers that will neither rupture on freezing nor leak upon thawing.
8.6 Storage and preservation
8.6.1 Samples shall be stored in such a way as to ensure that they are secure and suitably preserved for forensic examination.
8.6.2 Liquid samples and wet stains should generally be stored frozen (preferably at -20 degrees centigrade) unless they have otherwise been treated to render them stable for storage at an ambient temperature (for example, by drying or by use of a preservative or fixative)[footnote 3].
8.6.3 Unfrozen samples should be stored dry, at an ambient temperature and out of direct sunlight.
9. Case assessment and interpretation
9.1 Case assessment
9.1.1 The task-relevant (pedigree) information supplied by the authority submitting (or proposing to submit) the exhibits for relationship testing should be reviewed by the laboratory and propositions should be formulated accordingly.
9.1.2 In criminal cases, and when formulating the defence proposition or hypothesis (Hd), unless contrary information regarding a specific defence is received, the laboratory will be entitled to presume that the defendant will deny the alleged biological relationship. Any assumptions made shall be stated explicitly.
9.1.3 For body identification work and when formulating the Hd, the laboratory will be entitled to presume that the deceased is unrelated to the putative family of interest.
9.1.4 In addition, the laboratory should give due consideration as to whether the application of the autosomal short tandem repeat (STR) tests at their disposal could adequately address the issues in the case. See Annex 1, which considers the power of autosomal STR tests for addressing certain biological relationships and contains additional guidance.
9.1.5 If there is doubt as to whether the proposed tests can satisfactorily address the propositions, the laboratory shall inform the submitting authority of this. However, if the submitting authority nevertheless wishes to continue with testing, then this duty shall be considered to have been discharged.
9.1.6 In disaster victim identification cases where there are multiple fatalities and some within the same family, the laboratory shall alert the submitting authority that the tests employed may not be able to differentiate between close relatives involved in the same incident.
9.1.7 Where appropriate, the laboratory shall recommend the application of additional testing (such as Y-STRs or mitochondrial DNA) if these techniques offer a more suitable alternative to addressing the issue(s) in the case. This duty exists whether the forensic science provider (FSP) is capable of providing those services or not.
9.2 Interpretation and evaluation of evidence
9.2.1 After testing is complete, the genetic findings (i.e. the resulting DNA profiles) should be evaluated in order to determine the degree of support for both the prosecution proposition or hypothesis (Hp) and the Hd.
9.2.2 For those parent/child cases in which it is possible to formally exclude a biological relationship, the laboratory should establish and document their ‘exclusion’ criteria (see 9.10).
9.2.3 The statistical assessment of the weight of evidence in relationship cases is fundamentally different from ‘standard’ identification cases. The required formulae for some of the more common relationships are published in standard academic textbooks on the subject, see Buckleton et al., 2016.
9.2.4 However, for more complicated situations, bespoke formulae would need to be derived individually based on the specific pedigree and so the assistance of software is advisable (see section 9.3).
9.2.5 The basic process of evidential evaluation follows essentially the same framework as ‘standard’ identification cases by:
a. Formulating both prosecution and defence propositions based on the case information;
b. Assigning probabilities to those propositions using the data; and
c. Computing a likelihood ratio as a measure of the evidential weight of evidence.
9.2.6 The same UK allele frequency data (see the Home Office publication) [20] that is used for ‘standard work’ can be used for assignment of probabilities in relationship testing cases.
9.2.7 For the purposes of a calculation, a practitioner may elect to choose one allele frequency database (which is most similar to the biogeographic ancestry of the defendant) or perform multiple calculations using the different available frequency databases.
9.2.8 If multiple calculations are performed, the laboratory should document its policy and procedure for reporting the outcome(s).
9.2.9 Consideration of how to deal with ‘rare’ or ‘zero frequency’ alleles may be necessary in some cases.
9.2.10 An allowance for co-ancestry may be necessary in some cases.
9.2.11 An allowance for linked loci may be necessary in some cases.
9.2.12 An allowance for mutation may be necessary in some cases.
9.2.13 Further detailed guidance regarding the use of population databases and the making of various statistical allowances is provided in section 9.11.
9.3 General recommendations on the use of calculation software
9.3.1 Although manual calculations are possible for some of the simpler biological relationships, due to the large number of mathematical operations necessary and the error-prone nature of such calculations, it is strongly recommended that calculation software be employed to prevent mathematical errors.
9.3.2 All software applications require that genotypic information be entered and so it is recommended that, if possible, automatic import functionality be used to minimise the risk of transposition errors. Where allele data are keyed in manually, additional checks should be undertaken to ensure the accuracy of the input data.
9.3.3 Output files should be generated and stored (as either electronic copy or hard copy or both) so that these data can be inspected at a later date. The output should contain a record of the alternative prosecution and defence propositions considered and sufficient detail to enable checking, auditing and defence review. Where such a record is not part of the software output, manual records shall be made and retained.
9.3.4 It is recommended that the user interface should be intuitive and easy to use (for example, using a graphical user interface for ‘button and menu’ clicking rather than requiring entry of command lines).
9.3.5 It is recommended that the source code and embedded data files (such as allele frequency tables) should be locked to prevent users from inadvertently altering it.
9.3.6 Prior to any release of updates and/or new versions of calculation software a risk assessment should be carried out and, if appropriate, the new versions should be re-validated.
9.4 Recommendations regarding functionality for calculation software
9.4.1 It is recommended that any software application possesses the following calculation functionality.
a. The ability to use different geographic ancestry (population) groups.
b. The ability to apply some form of sampling adjustment to allele frequencies.
c. The ability to allow for co-ancestry/population substructure.
d. The ability to make some form of allowance for:
i. Germline mutation; and/or
ii. Linked loci.
9.5 Recommendations regarding the types of biological relationship that could be analysed by software
9.5.1 It is recommended that calculation software shall be designed to evaluate biological relationships. These include the following biological relationships:
a. Parent/child;
b. Siblings/half siblings;
c. Aunt/uncle;
d. Grandparents;
e. First cousins; and
f. Any of the above relationships involving incestuous pedigrees.
9.6 General guidance on the validation of statistical software tools
9.6.1 It is a general requirement that all software tools deployed by FSPs shall have been validated to a standard acceptable to the key stakeholders. Those designed for relationship testing applications are no different.
9.6.2 The International Society for Forensic Genetics (ISFG) has published guidelines for the validation of software performing bio-statistical calculations for forensic genetic calculations, see Gjertson et al., 2007. This sets out the minimum requirements for validation and covers both developmental and operational validation.
9.6.3 For the purposes of validation, the sections on ‘test methods and method validation’ and ‘estimation of uncertainty’ in the FSR codes shall apply. Refer to FSR-G-201 for more guidance on the necessary validation steps.
9.6.4 Whilst intended for the validation of mixtures interpretation software, the validation principles set out in FSR-G-223 shall also apply to commercial software or software developed ‘in house’ by FSPs for the purpose of relationship testing.
9.6.5 For calculation tools such as Excel spreadsheets the onus is on the party employing such a tool to validate it by demonstrating that the functions and calculations it performs are mathematically correct.
9.7 Dealing with apparent inconsistencies in the Mendelian inheritance pattern
9.7.1 Where necessary, the laboratory should take into account, statistically, the presence of a germline mutation. It is not permissible, when estimating the weight of evidence, to ignore a locus exhibiting an incompatibility with the Mendelian inheritance pattern.
9.7.2 The laboratory shall document its policy and procedure for dealing with apparent inconsistencies in the Mendelian inheritance pattern.
9.7.3 Mendelian inconsistencies can arise – even though two individuals are biologically parent and offspring – via the mechanism of mutation. When these mutations occur in the germline cells (i.e. at meiosis) the change becomes a heritable trait.
9.7.4 There are three types of germline mutational change that can potentially affect the results of relationship tests performed using STRs.
a. Base mutations and/or indels affecting a primer binding site can abolish the priming of one allele at a locus leading to the presence of a silent (null) allele that is not observable via a polymerase chain reaction. Hereafter this is referred to as a primer binding site mutation.
b. Alterations (for example, due to strand slippage or unequal crossover) to the number of repeats at a STR locus. Mostly (but not exclusively) these produce ‘single-step’ changes (i.e. the number of repeats either expands or contracts by one full repeat unit).
c. Gross chromosomal rearrangements (such as uniparental disomy [UPD] and aneuploidy).
9.7.5 Some of these phenomena can result in the biological offspring apparently having no allele in common with the biological mother (maternal mutation) and/or the biological father (paternal mutation), which is therefore inconsistent with the expected pattern of Mendelian inheritance given that the biological relationship is true.
9.7.6 Some germline mutations can occur without affecting the Mendelian inheritance pattern (so-called ‘covert’ mutations).
9.7.7 It is recommended that primer binding site mutations be resolved by further analysis using alternative primer sets. If this is not possible, or the further analysis fails to demonstrate the presence of an allele, then it is recommended that the locus be treated statistically as a germline mutation.
9.7.8 Suspected cases of UPD (based on the apparent homozygosity of the child at the locus) can be investigated by testing other markers located on the same section of chromosome, see Cavalheiro et al., 2020. If this is not possible, then it is recommended that the locus be treated statistically as a germline mutation.
9.7.9 Where it is necessary to allow statistically for the effects of an overt, repeat change mutation when computing the likelihood ratio (LR), implementation of a mutation model will be required. Several mutation models are available but the one most commonly used in the forensic community is the ‘stepwise’ mutation model, see Ohta and Kimura, 1973 [23]. In this model, alleles are deemed to mutate in an iterative stepped fashion (with multistep changes being less probable than single-step changes).
9.7.10 Mutation rates from collections of data have been published by the National Institute of Standards and Technology and can be used to derive or infer transition probabilities. Rates are known to be different for males and females.
9.7.11 In addition, in males the mutation rate appears to increase with chronological age, see Qin et al., 2015. This has been ascribed to the fact that in females, the oocytes are laid down at birth, whereas spermatozoa are made continuously in men and so, over time, have the propensity to amass more mutations.
9.7.12 It is recommended that calculation software contains the necessary functionality to include an allowance for mutational events.
9.8 Dealing with linked loci
9.8.1 For any autosomal multiplex used by the laboratory to investigate biological relationships, the laboratory should establish which (if any) of the loci are syntenic and which of those syntenic loci will require a formal linkage allowance.
9.8.2 The laboratory shall document this policy accordingly.
9.8.3 For those loci deemed to require it, the laboratory should incorporate an allowance for linkage into the reported LR.
9.8.4 The laboratory shall document its policy and procedure for dealing with linked loci (see 9.8.9 to 9.8.10).
9.8.5 STR loci located on the same chromosome are said to be ‘syntenic’. With the increasing size of multiplex kits, it is more likely that syntenic loci will be present. The corollary of this is that because the loci are not on separate chromosomes, they will exhibit linkage, therefore an allowance should be incorporated where necessary.
9.8.6 The closer together the loci are on the chromosome, the more tightly they will be linked. Some syntenic loci are so far apart on the chromosome that the effects of linkage will be negligible. For some more closely linked loci, the effects can be more significant.
9.8.7 Of the loci currently (2021) used for the UK National DNA Database®, only vWA and D12S391 are considered close enough (11cM) to warrant an allowance to be made.
9.8.8 Although genetic linkage has no impact on LR calculations in standard parentage cases, LRs can be substantially impact other relationships where the effects of linkage are not taken into account, see Tillmar and Phillips, 2017. Furthermore, linkage effects can result in an overestimation of the LR in scenarios involving incest, see O’Connor and Tillmar, 2012.
9.8.9 A formal statistical adjustment can be made using recombination frequencies, see Bright et al., 2014.
9.8.10 Alternatively, a simpler adjustment can be achieved by dropping one locus from each syntenic pair post-calculation. It is most conservative (i.e. most favourable to the defendant) to drop the locus with the highest calculated LR.
9.9 Making an allowance for co-ancestry (fixation index)
9.9.1 An allowance for co-ancestry should be applied to all calculations for criminal cases.
9.9.2 An allowance for co-ancestry is not required for body identifications or for those cases where the defendant is positively asserting (as opposed to denying) a biological relationship.
9.9.3 An allowance for population substructure can be made using a fixation index (Fst) adjustment (θ). This is in line with the policy for criminal cases involving DNA identification. This has been set at 0.03 (or 3%) and ensures that a calculation is not unfavourable towards a defendant who is taken to be disputing the biological relationship, see Steele and Balding, 2014 and Steele et al., 2014.
9.9.4 However, if a person on trial is positively asserting a biological relationship then θ may be set to zero (or at a much lower value than 0.03) at the discretion of the practitioner. This is because in such cases, any over reduction in evidential strength caused by applying a larger than required co- ancestry allowance operates against the interests of the defendant.
9.9.5 In body identification cases, there is no prima facie reason to skew the calculation in the favour of any particular party and so θ may be set at a more realistic value than 0.03 at the discretion of the practitioner.
9.10 Guidance on declaring an exclusion
9.10.1 In theory a LR can be calculated for any given pedigree (irrespective of how many mutational events might be required to explain the evidence under the Hp). If the LR was less than 1, the evidential strength could then be expressed in terms of the degree of support for the Hd. For each locus for which a mutation must be invoked under the Hp, the corresponding probability will be very low (often in the order of 0.001). When several loci are involved the LR will become very small very quickly.
9.10.2 In reality, therefore, and for pragmatic reasons, rather than report a large LR supporting the Hd a simple ‘exclusion’ is declared.
9.10.3 It follows logically from this analysis that if the pedigree is fully consistent with the Hp, a LR calculation should be undertaken. However, if there are inconsistencies under the Hp at ‘several’ loci, then an ‘exclusion’ may be declared instead of undertaking a calculation.
9.10.4 A laboratory policy should be set and guidance issued to practitioners as to how many loci must exhibit inconsistencies under the Hp in order to trigger a declaration of an exclusion.
9.10.5 The number of steps required for the proposed mutation(s) will have a bearing on this decision because, under the stepwise mutation model, multistep changes to the repeat units are inherently less probable than single-step repeat unit changes.
9.11 Guidance On Using Allele Frequency (Population) Databases
9.11.1 Allelic frequency (population) databases are required for statistical evaluations to be performed in all DNA cases. A population database is usually defined according to ethnic appearance (biogeographic ancestry group) and/or geographical descriptors.
9.11.2 In identification work, forensic providers in the UK make use of four different data collections, which represent most of the population of the UK, see Home Office publication [20]. These same data collections can be used for relationship testing purposes.
9.11.3 In the event that additional data collections are required, the following paragraphs provide recommendations as to their construction.
9.11.4 It is recommended that samples collected to form an allele frequency database should be samples drawn from a pre-defined group (usually defined ethno-geographically).
9.11.5 It is recommended that proportionate efforts should be made to check for, and exclude from the data set, relatives or duplicate samples.
9.11.6 It is recommended that at least 400 alleles (200 individuals) are surveyed. The effects of sampling variation will be more pronounced in databases containing small numbers of individuals and the rarer alleles may not always be detectable in a data sets of this size, see Gusmão et al., 2017.
9.11.7 In order to confirm that the database created is robust see Bodner et al., 2016 it is recommended that it be examined using a regime of statistical tests that meet the requirements of the international forensic science community.
9.11.8 Guidance can be found in FSR-G-213 Allele frequency databases and reporting guidance for the DNA (Short Tandem Repeat) profiling.
9.11.9 It is recommended that if possible, the data collected should be externally published or, alternatively, made available in a public archive so that it might be available to be scrutinised by others.
10. Reporting considerations
10.1 Reporting format and contents
10.1.1 The legal requirements as to the content and formatting of reports for use in the criminal justice system are documented elsewhere Legal Obligations FSR-1-400 and Expert Report Guidance FSR-G-200. The specific content of a report is only covered in this section insofar as it concerns the additional elements that are required for reporting the outcomes of investigations into biological relationships.
10.1.2 The following shall appear within a written report dealing with the investigation of biological relationships:
a. A summary of the pedigree information supplied and on which the analysis has been based;
i. A statement of the specific propositions that were used to compute the likelihood ratio (LR); and
b. A declaration of any assumptions that have been included within the analysis (for example, that a child’s maternity has been assumed to be true).
10.1.3 The LR shall always be reported.
10.1.4 The calculation of a posterior probability (or any other value that requires an assessment of the priors of a biological relationship) should not be reported.
10.1.5 In line with other policies, a LR of ‘one billion’ should be adopted as the upper reporting boundary for all relationship tests.
10.1.6 Forensic science providers may unilaterally decide to self-impose a lower testing boundary for certain types of relationship (for example, a policy to try to obtain a minimum LR). However, the test results obtained shall be disclosed to the submitting authority even if the required threshold is not reached, accompanied by an explanation.
10.1.7 Unless a specific defence is advanced, or the submitting authority otherwise requests it, there is no duty placed on the practitioner to unilaterally consider, explore and report on other scenarios involving individuals related to the defendant.
10.2 Ethical dilemmas and the Equality Act 2010
10.2.1 During DNA profiling tests information as to biological sex is obtained, which could contradict the accepted gender of a person in the pedigree. In addition, and in some cases, pedigree information regarding accepted (known) relationships is provided that, based on the DNA profiling results, can be demonstrated to be false (for example, the non-paternity of a child).
10.2.2 Primer binding site mutations and other mutations in DNA profiling can sometimes lead to a DNA profile from an individual whose sex is male presenting with an apparent X,X (female) genotype. Certain genetic anomalies can also present as multiple copies of X and Y in both males and females. Under normal circumstances it is not necessary to make reference to these situations in DNA profiling. However, certain case circumstances may lead to the testing of individuals who identify with a gender other than the one they were allocated at birth. Certain individuals may have completed the necessary legal steps to use a new legal name and be recognised as, for example, female. Transgender individuals are recognised as a protected group as defined by the Equality Act 2010. For the purposes of the Act, DNA testing laboratories are service providers. It is necessary for the scientist to consider the needs of the parties involved in the case and be respectful of their wishes when compiling reports. The various circumstances need not be defined here, but in general terms the following apply.
10.2.3 Use the names by which all parties wish to be identified.
10.2.4 Only make references to sex where absolutely necessary and be sure to recognise that there is a difference between sex and gender, whether a person has received a gender recognition certificate or not.
10.2.5 Contradictory sex and gender information can arise in the following different ways.
10.2.6 A genetic anomaly at the Amelogenin locus can result in the non-priming of the Y homolog, see Steinlechner et al., 2002. This produces an apparently female result even though the person is genetically male.
10.2.7 In very rare conditions (for example, Androgen Insensitivity Syndrome), a male lacks the hormonal receptors meaning that despite being genetically male he appears, phenotypically, to be female.
10.2.8 The person may identify with a gender other than the one they were allocated at birth.
10.2.9 Contradictory sex/gender information should be brought to the attention of the submitting authority as it might betray a sample mix-up prior to receipt at the laboratory. The information should never be disclosed to third parties, including directly to the donor(s) themselves. Whether, and how, to investigate such occurrences and/or decisions regarding disclosing the information to donors is a decision that should rest with the submitting authority.
10.2.10 When testing a foetus, the biological mother may not know the gender/sex of the child and might wish not to know this information. The foetal sex can be reported, and the relevant authority should consider whether they should not disclose this information.
10.2.11 If there are any genetic inconsistencies that would call into question known/given relationships in a given pedigree and which would undermine the analysis, the laboratory should disclose this to the submitting authority. The testing laboratory should not disclose such information to any third parties, including to the donors themselves. Whether, and how, to investigate such occurrences and/or decisions regarding disclosing the information to donors is a decision that should rest with the submitting authority.
10.2.12 For deceased individuals, it is recommended that any tissue samples be held by the DNA testing laboratory for the minimum amount of time possible (for example, only until a DNA profile has been prepared) and thereafter shall be returned promptly to the seizing authority.[footnote 4]
10.2.13 For products of conception, it is recommended any tissue samples be held by the DNA testing laboratory only until a DNA profile has been prepared and then be returned promptly to the seizing authority.[footnote 5]
10.2.14 Human tissue should always be handled in accordance with the provisions of the Human Tissue Act 2004.
11. Dealing with unusual features and genetic anomalies in DNA profiles
11.1.1 The majority of reference samples processed for relationship testing are expected to be single source with each locus having one or two alleles (depending on its heterozygosity). Where two alleles are present, and provided sufficient DNA is present, the allelic peaks should be balanced. Using neoplastic tissue as a surrogate sample can reveal gross chromosomal rearrangements and loss of whole chromosomes, see Budimlija et al., 2009.
11.1.2 Occasionally, however, samples exhibit reproducible aberrant di-allelic or tri- allelic patterns, see Clayton et al., 2004 and/or the sample appears to be reproducibly mixed despite being expected to be single source, see Rettner, 2016.
11.1.3 Aberrant allelic patterns are the results of genetic rearrangements (somatic mutations, copy number variation, trisomy and so on). These will only matter if the rearrangement affects the gametes and then causes an inconsistency the Mendelian inheritance pattern.
11.1.4 Additionally, they may present problems for software in subsequent calculations when a genotype has to be input. In such instances, and in order to use a software calculator, it is permissible to exclude such a locus from a calculation provided that the results at the locus are still compatible with the asserted biological relationship.
11.1.5 Unexpected mixtures obtained from the testing of a reference sample can be the result of a number of reasons (other than sample contamination).
11.1.6 If a personal effect (such as a toothbrush) has been tested as surrogate reference sample, then this could simply mean that another person has used the article concerned.
11.1.7 Samples taken from products of conception often produce mixed DNA profiles due to the presence of mixed maternal and foetal tissues. If a sample from the mother has been provided it may be possible to condition the mixture on the mother’s DNA profile, thereby determining the paternally inherited components in the DNA profile of the foetus. It is possible to compare these paternal DNA components to the DNA profile of any alleged father.
11.1.8 Occasionally, there can be biological reasons for the individual being tested to exhibit a mixed DNA profile. Such reasons include being the recipient of an allogenic bone marrow transplant, see Pope et al., 2006 or rare instances of human chimerism, Yunis et al., 2007.
11.1.9 All samples giving unexpected results should be duplicated prior to initiating further investigation. If multiple additional peaks are observed reproducibly within a DNA profile, it may be necessary to first consider and investigate whether the original donor sample could have been contaminated with exogenous DNA at the laboratory (or during the original sampling).
11.1.10 The next step is to request from the submitting authority a replacement sample for the individual in question. If the phenomenon persists then it is likely that the person is exhibiting an in vivo mixed DNA profile for one of the reasons outlined above.
11.1.11 Sometimes, targeting a different type of sample (for example, hair follicles rather than blood) can resolve the issue, see Chaudhary et al., 2015.
12. Acknowledgements
12.1.1 The Forensic Science Regulator would like to thank Cellmark Forensic Services, DNA Principal Forensic Services, Eurofins Forensic Services, Forensic Science Ireland, Forensic Science Northern Ireland,Key Forensic Services Ltd, the Scottish Police Authority, members of the Forensic Science Regulator’s DNA Specialist Group and the Forensic Science Regulation Unit (FSRU).
13. Review
13.1.1 This publication will be subjected to periodic review with a frequency as determined by the Forensic Science Regulator.
13.1.2 The Forensic Science Regulator welcomes comments regarding the content of this publication. Please send them to the address as set out at: www.gov.uk/government/organisations/forensic-science-regulator,
13.1.3 or please email them to: FSREnquiries@homeoffice.gov.uk
14. Abbreviations and Acronyms
cM
CentiMorgans
DNA
Deoxyribonucleic acid
FSI
Forensic Science International
FSP
Forensic science providers
FSR
Forensic Science Regulator
FST
Fixation index
Hd
The defence proposition or hypothesis
Hp
The prosecution proposition or hypothesis
ISFG
International Society for Forensic Genetics
LR
Likelihood ratio
NDNAD
National DNA database®
NIST
National Institute of Standards and Technology
PACE
Police and Criminal Evidence Act 1984
SNP
Single nucleotide polymorphism
STR
Short tandem repeat
15. References
British Standard, BS EN ISO/IEC 17025, ‘General requirements for the competence of testing and calibration laboratories’.
International Laboratory Accreditation Cooperation, ILAC G19:08/2014 ‘Modules in a Forensic Science Process’, [online]. [Accessed 10 03 2021].
Forensic Science Regulator, ‘Codes of Practice and Conduct for Forensic Science Providers and Practitioners in the Criminal Justice System,’ FSR-C- 100, [online]. [Accessed 10 03 2021].
Forensic Science Regulator, ‘DNA Analysis,’ FSR-C-108, [online]. [Accessed 10 03 2021].
Forensic Science Regulator ‘Validation,’ FSR-G-201, [online]. [Accessed 10 03 2021].
Forensic Science Regulator, ‘The interpretation of DNA evidence (including low-template DNA),’ FSR-G-202, [online]. [Accessed 14 03 2021].
Forensic Science Regulator, ‘The control and avoidance of contamination in laboratory activities, involving DNA evidence recovery and analysis,’ FSR-G- 208, [online]. [Accessed 10 03 2021].
Forensic Science Regulator, ‘Allele frequency databases and reporting guidance for the DNA (short tandem repeat) profiling,’ FSR-G-213, [online]. [Accessed 10 03 2021].
Forensic Science Regulator, ‘DNA mixture interpretation,’ FSR-G-222, [online]. [Accessed 10 03 2021].
Forensic Science Regulator, ‘Software validation for DNA mixture interpretation,’ FSR-G-223, [online]. [Accessed 10 03 2021].
Forensic Science Regulator, ‘DNA contamination detection: The management and use of staff elimination DNA databases,’ FSR-P-302, [online]. [Accessed 10 03 2021].
Police and Criminal Evidence Act 1984 [online]. [Accessed 19 03 2021].
Home Office Circular 1/2006, ‘The Application for Access to a DNA Profile for Paternity’, [online]. [Accessed 19 03 2021].
Faculty of Forensic and Legal Medicine, ‘Guidance on Paternity Testing’, [online]. [Accessed 19 03 2021].
Interpol, Disaster Victim Identification (DVI) Protocols and Standards, [online]. [Accessed 19 03 2021].
Interpol, Best Practice Principles: Recommendations on the Use of DNA for the Identification of Missing Persons and Unidentified Human Remains, [online]. [Accessed 19 03 2021].
College of Policing, Authorised Professional Practice Disaster Victim Identification, [online]. [Accessed 14 03 2021].
Prinz, M., Carracedo, A., Mayr, W. R., Morling, N., Parsons, T. J., Sajantila, A., Scheithauer, R., Schmitter, H. and Schneider P. M. (2007) ‘DNA Commission of the International Society for Forensic Genetics (ISFG): Recommendations regarding the role of forensic genetics for disaster victim identification (DVI)’, Forensic Science International: Genetics, vol. 1 (1), pp 3–12. doi: 10.1016/j.fsigen.2006.10.003.
Buckleton, J. S., Bright, J.-A. and Taylor, D. (2016) ‘Forensic DNA Evidence Interpretation’. Boca Raton, FL: CRC Press.
Home Office, ‘Data to support the implementation of national DNA database DNA-7 profiling’, [online]. [Accessed 19 03 2021].
Gjertson, D. W., Brenner, C. H., Baur, M. P., Carracedo, A., Guidet, F., Luque, J. A., Lessig, R., Mayr, W. R., Pascali, V. L., Prinz, M., Schneider, P. M. and Morling, N. (2007) ‘ISFG: Recommendations on biostatistics in paternity testing’, Forensic Science International: Genetics, vol. 1 (3), pp 223–231. doi: 10.1016/j.fsigen.2007.06.006.
Cavalheiro, C. P., Avila, E., Gastaldo, A. Z., Graebin, P., Motta, C. H. A., Rodenbusch, R. and Alho, C. S. (2020) ‘Uniparental disomy of chromosome 21: A statistical approach and application in paternity tests’, Forensic Science International: Genetics, vol. 49, p. 102368. doi: org/10.1016/j.fsigen.2020.102368.
Ohta, T. and Kimura, M. (1973) ‘A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population’, Genet. Res., vol. 22, pp 201–204.
National Institute of Standards and Technology, Apparent Mutations Observed at STR Loci in the Course of Paternity Testing, [online]. [Accessed 19 03 2021].
Qin, X.-Q., Yin, C.-Y., Ji, Q., Li, K., Fan, H.-T., Yu, Y.-F., Bu, F.-L., Hu, L.-L., Wang, J.-W., Mu, H.-F., Haigh, S. and Chen, F. (2015) ‘Mutation rate analysis at 19 autosomal microsatellites’, Electrophoresis, vol. 36, pp 1633– 1639.
Tillmar, A. O. and Phillips, C. (2017) ‘Evaluation of the impact of genetic linkage in forensic identity and relationship testing for expanded DNA marker sets’, Forensic Science International: Genetics, vol. 26, pp 58–65.
O’Connor, K. L. and Tillmar, A. O. (2012) ‘Effect of linkage between vWA and D12S391 in kinship analysis’, Forensic Science International: Genetics, vol. 6, no. 6, pp 840–844.
Bright, J., Hopwood, A., Curran, J. M. and Buckleton, J. S. (2014) ‘A Guide to Forensic DNA Interpretation and Linkage’, [online]. [Accessed 19 03 2021].
Steele, C. and Balding, D. (2014) ‘Choice of population database for forensic DNA profile analysis’, Science and Justice, vol. 54, no. 6, pp 487–493.
Steele, C., Syndercombe-Court, D. and Balding, D. (2014) ‘Worldwide Fst estimates relative to five continental-scale populations’, Annals of Human Genetics, vol. 78, pp 468–477.
Gusmão, L., Butler, J. M., Linacre, A., Parson, W., Roewer, L., Schneider, P. M. and Carracedo, A. (2017) ‘Revised guidelines for the publication of genetic population data’, Forensic Science International: Genetics, vol. 30, September, pp 160–163. doi: 10.1016/j.fsigen.2017.06.007.
Bodner, M., Bastisch, I., Butler, J. M., Fimmers, R., Gill, P., Gusmão, L., Morling, N., Phillips, C., Prinz, M., Schneider, P. M. and Parson, W. (2016) ‘Recommendations of the DNA Commission of the International Society for Forensic Genetics (ISFG) on quality control of autosomal Short Tandem Repeat allele frequency databasing (STRidER)’, Forensic Science International: Genetics, vol. 24, pp 97–102.
Forensic Science Regulator, ‘Legal Obligations for Witnesses Providing Expert Evidence,’ FSR-I-400, [online]. [Accessed 19 03 2021].
Forensic Science Regulator, ‘Expert Report Guidance,’ FSR-G-200, [online]. [Accessed 19 03 2021].
The Equality Act 2010, [online]. [Accessed 10 03 2021].
Steinlechner, M., Berger, B., Niederstätter, H. and Parson, W. (2002) ‘ Rare failures in the amelogenin sex test’, International Journal of Legal Medicine, vol. 116, pp 117–120.
Human Tissue Act 2004, [online]. [Accessed 19 03 2021].
Budimlija, Z., Lu, C., Axler-DiPerte, G., Seifarth, J., Popiolek, D., Fogt, F. and Prinz, M. (2009) ‘Malignant tumors and forensics-dilemmas and proposals’, Croat. Med. J., vol. 50 (3), June, pp 218–227.
Clayton, T. M., Guest, J. L., Urquhart, A. J. and Gill, P. D. (2004) ‘A genetic basis for anomalous band patterns encountered during DNA STR profiling’, Journal Forensic Science, vol. 49 (6), November, pp 1207–1214.
Rettner, R. (2016) ‘3 Human Chimeras That Already Exist’, LiveScience, August 8, 2016, [online]. [Accessed 19 03 2021].
Pope, S., Chapman, H. and Lambert, J. (2006) ‘The Effect of Bone Marrow Transplants on DNA profiles; a case example’, Science and Justice, vol. 46, no. 4, pp 231–237.
Yunis, E. J., Zuniga, J. and Romero, V. (2007) ‘Chimerism and tetragametic chimerism in humans: implications in autoimmunity, allorecognition and tolerance’, Immunol. Res., vol. 38 (1–3), pp 213–236. doi: 10.1007/s12026- 007-0013-3.
Chaudhary, G., Dogra, T. and Raina, A. (2015) ‘Evaluation of blood, buccal swabs, and hair follicles for DNA profiling technique using STR markers’, Croatian Medical Journal, vol. 56, pp 239–245. doi: 10.3325/cmj.2015.56.239.
Association of Forensic Science Providers (2009) ‘Standards for the formulation of evaluative forensic science expert opinion’, Science and Justice, vol. 49, pp 161–164. doi: 10.1016/j.scijus.2009.07.004.
16. Further Reading
Balding, D. J. (1995) ‘Estimating products in forensic identification using DNA profiles’, J. Am. Stat. Assoc., 90, pp 839–844.
Balding, D. J. and Nichols, R. A. (1994) ‘DNA profile match probability calculation: how to allow for population stratification, relatedness, database selection and single bands’, Forensic Science International, 64, pp 125–140.
Bright, J., Curran, J. M. and Buckleton, J. S. (2013) ‘Relatedness calculations for linked loci incorporating subpopulation effects’, Forensic Science International: Genetics, 7, pp 380–383.
Bright, J., Curran, J. M., Hopwood, A. J., Puch-Solis, R. and Buckleton, J. S. (2013) ‘Consideration of the probative value of single donor 15-plex STR profiles in UK populations and its presentation in UK courts II’, Science and Justice, 53, p 371.
Budowle, B., Ge, J., Chakraborty, J., Eisenberg, A. J., Green, R., Mulero, J., Lagace, R. and Hennessy, L. (2011) ‘Population genetic analyses of the NGM STR loci’, International Journal of Legal Medicine, 125, pp 101–109. doi: 10.1007/s00414-010-0516-7.
Curran, J. M., Buckleton, J. S., Triggs, C. M. and Weir, B. S. (2002) ‘Assessing uncertainty in DNA evidence caused by sampling effects’, Science and Justice, 42, pp 29–37.
European Network of Forensic Science Institutes (ENFSI) (2014) Guidance on the Conduct of Proficiency Tests and Collaborative Exercises within ENFSI, [online]. [Accessed 19 03 2021].
Evett, I. W. and Weir, B. S. (1998) ‘Interpreting DNA evidence: Statistical genetics for forensic scientists’. Sinaur Associates Inc, ISBN 0 87893 155 4.
Forensic Science Regulator, ‘Validation – Use of casework material validation’, FSR-P-300. [online]. [Accessed 19 03 2021].
Gill, P., Phillips, C., McGovern, C., Bright, J. and Buckleton, J. (2012) ‘An evaluation of potential allelic association between the STRs vWA and D12S391: Implications in criminal casework and applications to short pedigrees’, Forensic Science International: Genetics, vol. 6, pp 477–486.
Hopwood, A. J., Puch-Solis, R., Tucker, V. C., Curran, J. M., Skerrett, J., Pope, S. and Tully, G. (2012) ‘Consideration of the probative value of single donor 15-plex STR profiles in UK populations and its presentation in UK courts’, Science and Justice, 52, pp 185–190.
The Royal Society and the Royal Society of Edinburgh (2017) ‘Forensic DNA Analysis – A primer for courts’ [online]. [Accessed 19 03 2021].
The Royal Society & The Royal Society of Edinburgh (2020) ‘The use of statistics in legal proceedings: a primer for courts’, [online]. [Accessed 26 03 2021].
United Kingdom Accreditation Service (2020) Policy on Participation in Proficiency Testing, TPS 47, edition 4, issued February 2020, [online]. [Accessed 19 03 2021].
Annex 1
17. Additional guidance for Addressing Certain Biological Relationships
17.1 The ‘power’ of STR kits for investigating specific biological relationships
17.1.1 In general, the power of any particular multiplex (or combination of multiplexes) in addressing a specific relationship will depend on the number and the heterozygosity of the loci employed. The more loci that are present and the more discriminating those loci are, the greater will be the power of the tests.
17.1.2 However, of much greater impact is the type of biological relationship being investigated and the number of other ‘known’ samples in the pedigree. So for instance, in a paternity test, the test will generally have more power if the non-questioned mother is also included in the testing rather than being absent (i.e. if a ‘trio’ is tested rather than a ‘duo’).
17.1.3 The greater the genetic distance between the parties, the less powerful the tests will be. So, tests of first-degree relationships (such as paternity and siblingship) are more powerful than tests of second-degree relationships (such as grandpaternity tests or avuncular – uncle/nephew or uncle/niece – tests).
17.1.4 Use of different minimum likelihood ratios (LRs) can provide useful information as to the expectations when carrying out tests of different relationships.
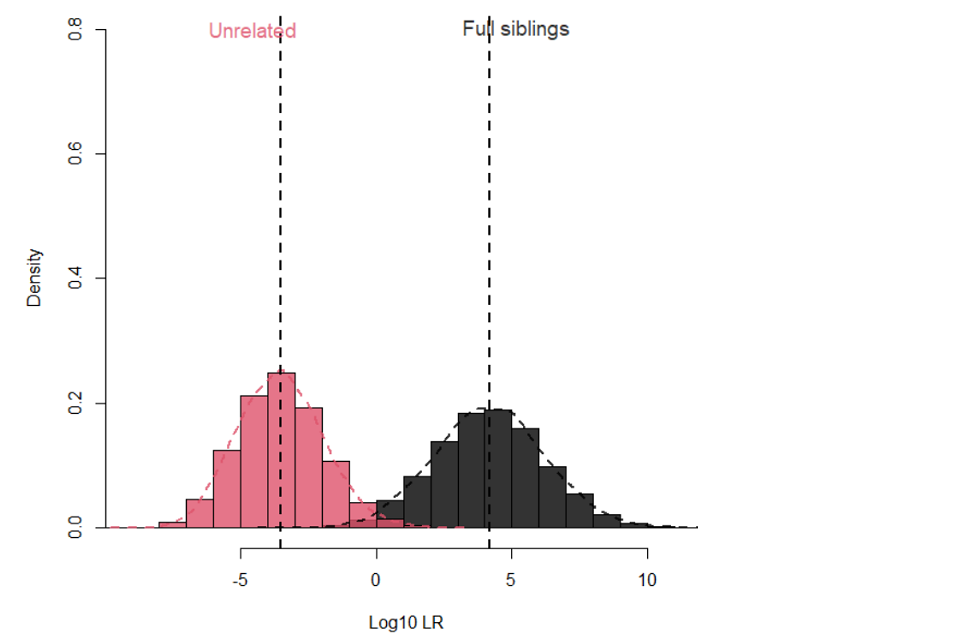
17.1.5 For example, using the 16 loci in DNA17 and a fixation index (Fst) of 0.03, about 87% of true full siblings will provide a LR greater than 100 with a 0.05% false positive rate. Using a standard descriptor of ‘moderate’ evidence does not reflect this state and it would seem more appropriate to describe such evidence as ‘strong, relative to the proposition’. Ninety three percent will provide a LR greater than 20 with a 0.2% false positive rate achieving a LR of 20, providing ‘moderately strong evidence, relative to the proposition’ might be proposed as providing a useful result. The false positive rate in this sense is the proportion of unrelated pairs that would provide a LR greater than the respective cut offs of a LR of 100 or 20, respectively.
17.1.6 Making use of additional loci, such the additional loci provided in Globalfiler™, or PowerPlex® Fusion, can improve the separation further. In this case 93% of true full siblings will provide a LR greater than 100 with a 0.05% false positive rate and 97.6% will achieve a LR greater than 20, on average, with a 0.1% false positive rate.
17.1.7 Figure 1 illustrates the separation between full siblings and unrelated pairs when using the 16 National DNA Database® (NDNAD) loci [20] and applying an Fst of 0.03 for Caucasians.
17.1.8
Figure 1: Separation between full siblings and unrelated pairs for 16 loci with Fst 0.03 for Caucasians
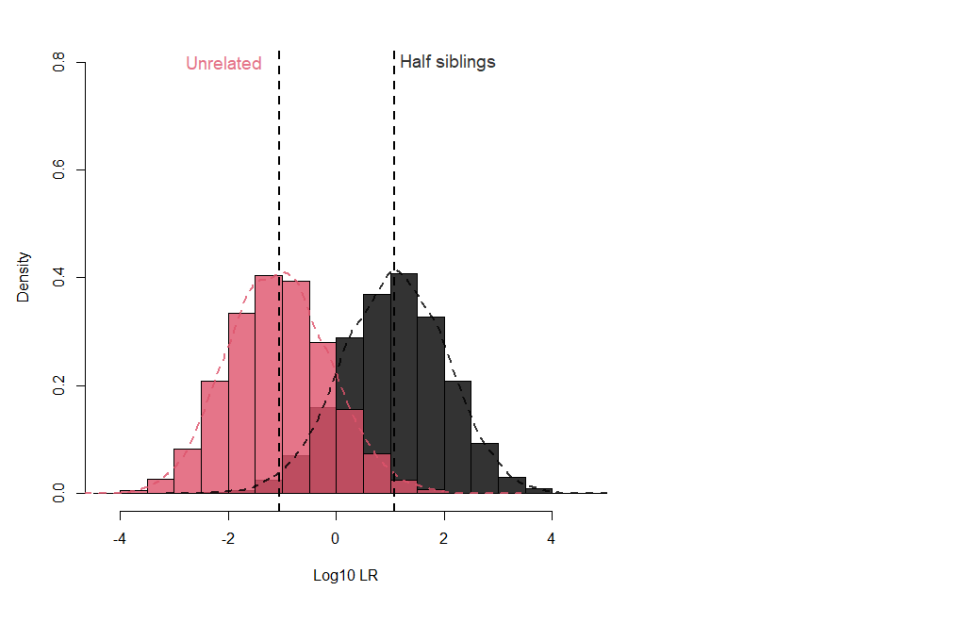
17.1.9 In contrast, when considering half siblings (or the genetically distanced equivalent grandparent-grandchild, or uncle/aunt-nephew/niece), only about 17% of true half siblings will provide a LR greater than 100 with a 0.1% false positive rate, and this only increases to about 40% with a LR greater than 20 and a false positive rate of 0.6%. Adding the Globalfiler™ or PowerPlex® Fusion loci increases these proportions to 33% (0.1% false positive) and 55% (0.8% false positive) respectively.
17.1.10 Additional loci should be used to improve confidence of a second-degree relationship if a LR greater than 100 is not achieved.
17.1.11 Figure 2 illustrates the poor separation between half siblings and unrelated pairs when using when using the 16 NDNAD loci and applying an Fst of 0.03 for Caucasians.
Figure 2: Separation between half siblings and unrelated pairs for 16 loci with Fst 0.03 for Caucasians
17.2 Testing of second- and third-degree pedigrees
17.2.1 The relationship testing of second-degree pedigrees will often lead to low or modest LRs. Adding ‘known’ relatives into such a pedigree is likely to improve the power of the test and may produce more meaningful results.
17.2.2 Testing third-degree pedigrees (without any other individuals in the pedigree) is unlikely to be satisfactorily addressed using standard autosomal short tandem repeat (STR) testing and LRs obtained are unlikely to be meaningful even where multiplex kits of 20+ loci are employed.
17.2.3 For second- and third-degree relationships, strong consideration should therefore be given to applying other supplementary DNA profiling techniques to investigate the relationship or to supplement the STR analyses.
17.2.4 Tests using uniparental markers and/or large single nucleotide polymorphism (SNPs) panels and/or large panels of sequenced STRs will have much greater investigative capacity for more distant relationships.
17.2.5 Research has shown that using a minimum 44 autosomal STR loci can satisfactorily address up to 50% of cousin/cousin relationships, 50% of true cousins give a LR greater than 10 with a 1% false error rate; (35% give a LR greater than 20 (0.7% error) and 15% give a LR greater than 100 (0.1% error).[footnote 6]
17.2.6 Given the information contained within this annex, it is not recommended to use a verbal scale, such as that published by the Association of Forensic Science Providers [44] for non-parentage relationships unless qualified due to nature of inheritance in the non-parentage pedigrees.
Published by:
The Forensic Science Regulator
5 St Philip’s Place
Colmore Row
Birmingham
B3 2PW
www.gov.uk/government/organisations/forensic-science-regulator
-
March 2021: England & Wales PACE [12] DNA Sampling Kit – Elimination Product Code: G00102- 25 ↩
-
March 2021:England & Wales PACE DNA Sampling Kit – Suspect Product Code: G00101-25 ↩
-
It should be noted that the use of preservatives such as formalin and formaldehyde have a very deleterious effect on DNA and can negatively impact downstream DNA processes. ↩
-
A sample of lysed cellular DNA may be retained by the laboratory to facilitate further testing or defence review. ↩
-
A sample of lysed cellular DNA may be retained by the laboratory to facilitate further testing or defence review. ↩
-
Personal communication Syndercombe-Court, D. Kings College London ↩

